Intellau_Celistic
5'3 KHHV Mentalcel
-
- Joined
- Aug 26, 2021
- Posts
- 166,411
Good frequencies for our use.
In the end only North Korea stays safe from the crackers.
Nope, they worship Soviet culture there and hate their own culture.
Bro, the people that you're whiteworshipping for, many of them don't care about you lol. This behaviour is cringeThis is why I am an antinatalist as a blackcel. Knowing that I am genetically, socially, economically and historically inferior to pretty much all races made me choose to never reproduce. I could never create an ethnic to get mogged in white society its too much for me already to be a subhuman.
Bro, the people that you're whiteworshipling for, many of them don't care about you lol. This behaviour is cringe
Bro, the people that you're whiteworshipping for, many of them don't care about you lol. This behaviour is cringe
| 1000Genomes_30x | Global | Study-wide | 6404 | G=0.6699 | A=0.3301 |
| 1000Genomes_30x | African | Sub | 1786 | G=0.9015 | A=0.0985 |
| 1000Genomes_30x | Europe | Sub | 1266 | G=0.5869 | A=0.4131 |
| 1000Genomes_30x | South Asian | Sub | 1202 | G=0.5782 | A=0.4218 |
| 1000Genomes_30x | East Asian | Sub | 1170 | G=0.5197 | A=0.4803 |
| 1000Genomes_30x | American | Sub | 980 | G=0.647 | A=0.353 |
| 1000Genomes_30x | Global | Study-wide | 6404 | G=0.6348 | A=0.3652 |
| 1000Genomes_30x | African | Sub | 1786 | G=0.8331 | A=0.1669 |
| 1000Genomes_30x | Europe | Sub | 1266 | G=0.5671 | A=0.4329 |
| 1000Genomes_30x | South Asian | Sub | 1202 | G=0.5790 | A=0.4210 |
| 1000Genomes_30x | East Asian | Sub | 1170 | G=0.6231 | A=0.3769 |
| 1000Genomes_30x | American | Sub | 980 | G=0.443 | A=0.557 |
| 1000Genomes_30x | Global | Study-wide | 6404 | G=0.8262 | A=0.1738 |
| 1000Genomes_30x | African | Sub | 1786 | G=0.9457 | A=0.0543 |
| 1000Genomes_30x | Europe | Sub | 1266 | G=0.7844 | A=0.2156 |
| 1000Genomes_30x | South Asian | Sub | 1202 | G=0.7479 | A=0.2521 |
| 1000Genomes_30x | East Asian | Sub | 1170 | G=0.7026 | A=0.2974 |
| 1000Genomes_30x | American | Sub | 980 | G=0.906 | A=0.094 |
| 1000Genomes_30x | Global | Study-wide | 6404 | T=0.6366 | G=0.3634 |
| 1000Genomes_30x | African | Sub | 1786 | T=0.5935 | G=0.4065 |
| 1000Genomes_30x | Europe | Sub | 1266 | T=0.6398 | G=0.3602 |
| 1000Genomes_30x | South Asian | Sub | 1202 | T=0.7429 | G=0.2571 |
| 1000Genomes_30x | East Asian | Sub | 1170 | T=0.5778 | G=0.4222 |
| 1000Genomes_30x | American | Sub | 980 | T=0.651 | G=0.349 |
| 1000Genomes_30x | Global | Study-wide | 6404 | A=0.5006 | G=0.4994 |
| 1000Genomes_30x | African | Sub | 1786 | A=0.7133 | G=0.2867 |
| 1000Genomes_30x | Europe | Sub | 1266 | A=0.5885 | G=0.4115 |
| 1000Genomes_30x | South Asian | Sub | 1202 | A=0.2912 | G=0.7088 |
| 1000Genomes_30x | East Asian | Sub | 1170 | A=0.2769 | G=0.7231 |
| 1000Genomes_30x | American | Sub | 980 | A=0.523 | G=0.477 |
| 1000Genomes_30x | Global | Study-wide | 6404 | C=0.5022 | T=0.4978 |
| 1000Genomes_30x | African | Sub | 1786 | C=0.7480 | T=0.2520 |
| 1000Genomes_30x | Europe | Sub | 1266 | C=0.4265 | T=0.5735 |
| 1000Genomes_30x | South Asian | Sub | 1202 | C=0.3577 | T=0.6423 |
| 1000Genomes_30x | East Asian | Sub | 1170 | C=0.4368 | T=0.5632 |
| 1000Genomes_30x | American | Sub | 980 | C=0.407 | T=0.593 |
| 1000Genomes_30x | Global | Study-wide | 6404 | A=0.2403 | G=0.7597 |
| 1000Genomes_30x | African | Sub | 1786 | A=0.1445 | G=0.8555 |
| 1000Genomes_30x | Europe | Sub | 1266 | A=0.4423 | G=0.5577 |
| 1000Genomes_30x | South Asian | Sub | 1202 | A=0.3136 | G=0.6864 |
| 1000Genomes_30x | East Asian | Sub | 1170 | A=0.0752 | G=0.9248 |
| 1000Genomes_30x | American | Sub | 980 | A=0.261 | G=0.739 |
| 1000Genomes_30x | Global | Study-wide | 6404 | A=0.6157 | G=0.3843 |
| 1000Genomes_30x | African | Sub | 1786 | A=0.4698 | G=0.5302 |
| 1000Genomes_30x | Europe | Sub | 1266 | A=0.4313 | G=0.5687 |
| 1000Genomes_30x | South Asian | Sub | 1202 | A=0.7055 | G=0.2945 |
| 1000Genomes_30x | East Asian | Sub | 1170 | A=0.9333 | G=0.0667 |
| 1000Genomes_30x | American | Sub | 980 | A=0.631 | G=0.369 |
| 1000Genomes_30x | Global | Study-wide | 6404 | T=0.9491 | C=0.0509 |
| 1000Genomes_30x | African | Sub | 1786 | T=0.9401 | C=0.0599 |
| 1000Genomes_30x | Europe | Sub | 1266 | T=0.9084 | C=0.0916 |
| 1000Genomes_30x | South Asian | Sub | 1202 | T=0.9767 | C=0.0233 |
| 1000Genomes_30x | East Asian | Sub | 1170 | T=1.0000 | C=0.0000 |
| 1000Genomes_30x | American | Sub | 980 | T=0.923 | C=0.077 |
| 1000Genomes_30x | Global | Study-wide | 6404 | T=0.6891 | C=0.3106, G=0.0003 |
| 1000Genomes_30x | African | Sub | 1786 | T=0.3001 | C=0.6999, G=0.0000 |
| 1000Genomes_30x | Europe | Sub | 1266 | T=0.6888 | C=0.3112, G=0.0000 |
| 1000Genomes_30x | South Asian | Sub | 1202 | T=0.9160 | C=0.0840, G=0.0000 |
| 1000Genomes_30x | East Asian | Sub | 1170 | T=0.9974 | C=0.0009, G=0.0017 |
| 1000Genomes_30x | American | Sub | 980 | T=0.752 | C=0.248, G=0.000 |
| 1000Genomes_30x | Global | Study-wide | 6404 | T=0.9500 | G=0.0500 |
| 1000Genomes_30x | African | Sub | 1786 | T=0.9916 | G=0.0084 |
| 1000Genomes_30x | Europe | Sub | 1266 | T=0.8555 | G=0.1445 |
| 1000Genomes_30x | South Asian | Sub | 1202 | T=0.9642 | G=0.0358 |
| 1000Genomes_30x | East Asian | Sub | 1170 | T=0.9991 | G=0.0009 |
| 1000Genomes_30x | American | Sub | 980 | T=0.920 | G=0.080 |
| 1000Genomes_30x | Global | Study-wide | 6404 | C=0.4605 | T=0.5395 |
| 1000Genomes_30x | African | Sub | 1786 | C=0.4093 | T=0.5907 |
| 1000Genomes_30x | Europe | Sub | 1266 | C=0.4179 | T=0.5821 |
| 1000Genomes_30x | South Asian | Sub | 1202 | C=0.4559 | T=0.5441 |
| 1000Genomes_30x | East Asian | Sub | 1170 | C=0.5735 | T=0.4265 |
| 1000Genomes_30x | American | Sub | 980 | C=0.480 | T=0.520 |
| 1000Genomes_30x | Global | Study-wide | 6404 | G=0.7313 | T=0.2687 |
| 1000Genomes_30x | African | Sub | 1786 | G=0.8303 | T=0.1697 |
| 1000Genomes_30x | Europe | Sub | 1266 | G=0.8104 | T=0.1896 |
| 1000Genomes_30x | South Asian | Sub | 1202 | G=0.7180 | T=0.2820 |
| 1000Genomes_30x | East Asian | Sub | 1170 | G=0.4265 | T=0.5735 |
| 1000Genomes_30x | American | Sub | 980 | G=0.829 | T=0.171 |
| 1000Genomes_30x | Global | Study-wide | 6404 | T=0.5448 | C=0.4552 |
| 1000Genomes_30x | African | Sub | 1786 | T=0.2990 | C=0.7010 |
| 1000Genomes_30x | Europe | Sub | 1266 | T=0.6051 | C=0.3949 |
| 1000Genomes_30x | South Asian | Sub | 1202 | T=0.8028 | C=0.1972 |
| 1000Genomes_30x | East Asian | Sub | 1170 | T=0.4231 | C=0.5769 |
| 1000Genomes_30x | American | Sub | 980 | T=0.744 | C=0.256 |
| gnomAD - Genomes | European | Sub | 75798 | G=0.83228 | A=0.16772 |
| gnomAD - Genomes | African | Sub | 41766 | G=0.34202 | A=0.65798 |
| gnomAD - Genomes | American | Sub | 13598 | G=0.82233 | A=0.17767 |
| gnomAD - Genomes | Ashkenazi Jewish | Sub | 3324 | G=0.8135 | A=0.1865 |
| Total | Global | 7920 | G=0.3301 | A=0.0000, C=0.6699, T=0.0000 |
| European | Sub | 7116 | G=0.2919 | A=0.0000, C=0.7081, T=0.0000 |
| African | Sub | 510 | G=0.820 | A=0.000, C=0.180, T=0.000 |
| African Others | Sub | 22 | G=0.77 | A=0.00, C=0.23, T=0.00 |
| African American | Sub | 488 | G=0.822 | A=0.000, C=0.178, T=0.000 |
| Asian | Sub | 4 | G=0.0 | A=0.0, C=1.0, T=0.0 |
| East Asian | Sub | 2 | G=0.0 | A=0.0, C=1.0, T=0.0 |
They have their own ideology. They don't even know Marx or Lenin.Nope, they worship Soviet culture there and hate their own culture.
They hate us because they can't be us.View attachment 548525
View attachment 548526
View attachment 548527
View attachment 548530
View attachment 548529
View attachment 548533
View attachment 548535
View attachment 548538
.....
.....
.....
......
View attachment 548536
View attachment 548540
View attachment 548541
View attachment 548537
They hate us because they can't be us.
| 1000Genomes_30x | Global | Study-wide | 6404 | A=0.8517 | G=0.1483 |
| 1000Genomes_30x | African | Sub | 1786 | A=0.7822 | G=0.2178 |
| 1000Genomes_30x | Europe | Sub | 1266 | A=0.8065 | G=0.1935 |
| 1000Genomes_30x | South Asian | Sub | 1202 | A=0.8819 | G=0.1181 |
| 1000Genomes_30x | East Asian | Sub | 1170 | A=0.9342 | G=0.0658 |
| European | Sub | 24732 | C=0.97016 | T=0.02984 |
| African | Sub | 3168 | C=0.9268 | T=0.0732 |
| African Others | Sub | 124 | C=0.895 | T=0.105 |
| African American | Sub | 3044 | C=0.9281 | T=0.0719 |
| European | Sub | 24368 | C=0.55052 | G=0.00000, T=0.44948 |
| African | Sub | 5610 | C=0.7626 | G=0.0000, T=0.2374 |
| African Others | Sub | 202 | C=0.861 | G=0.000, T=0.139 |
| African American | Sub | 5408 | C=0.7589 | G=0.0000, T=0.2411 |
| European | Sub | 14286 | G=0.42804 | A=0.57196 |
| African | Sub | 2946 | G=0.2648 | A=0.7352 |
| African Others | Sub | 114 | G=0.237 | A=0.763 |
| African American | Sub | 2832 | G=0.2659 | A=0.7341 |
| European | Sub | 18210 | G=0.53624 | A=0.46376 |
| African | Sub | 6746 | G=0.8663 | A=0.1337 |
| African Others | Sub | 232 | G=0.931 | A=0.069 |
| African American | Sub | 6514 | G=0.8640 | A=0.1360 |
| European | Sub | 7428 | C=0.3953 | A=0.0000, T=0.6047 |
| African | Sub | 530 | C=0.840 | A=0.000, T=0.160 |
| African Others | Sub | 20 | C=0.85 | A=0.00, T=0.15 |
| African American | Sub | 510 | C=0.839 | A=0.000, T=0.161 |
| European | Sub | 14286 | C=0.19320 | T=0.80680 |
| African | Sub | 2946 | C=0.3367 | T=0.6633 |
| African Others | Sub | 114 | C=0.447 | T=0.553 |
| African American | Sub | 2832 | C=0.3323 | T=0.6677 |
| 1000Genomes_30x | Global | Study-wide | 6404 | G=0.7281 | A=0.2719 |
| 1000Genomes_30x | African | Sub | 1786 | G=0.8903 | A=0.1097 |
| 1000Genomes_30x | Europe | Sub | 1266 | G=0.7844 | A=0.2156 |
| 1000Genomes_30x | South Asian | Sub | 1202 | G=0.6572 | A=0.3428 |
| 1000Genomes_30x | East Asian | Sub | 1170 | G=0.5453 | A=0.4547 |
| 1000Genomes_30x | American | Sub | 980 | G=0.665 | A=0.335 |
| 1000Genomes_30x | Global | Study-wide | 6404 | G=0.4811 | A=0.0016, C=0.5173 |
| 1000Genomes_30x | African | Sub | 1786 | G=0.8863 | A=0.0056, C=0.1081 |
| 1000Genomes_30x | Europe | Sub | 1266 | G=0.5245 | A=0.0000, C=0.4755 |
| 1000Genomes_30x | Global | Study-wide | 6404 | T=0.3734 | C=0.6266 |
| 1000Genomes_30x | African | Sub | 1786 | T=0.3371 | C=0.6629 |
| 1000Genomes_30x | Europe | Sub | 1266 | T=0.4573 | C=0.5427 |
| 1000Genomes_30x | South Asian | Sub | 1202 | T=0.5466 | C=0.4534 |
| 1000Genomes_30x | East Asian | Sub | 1170 | T=0.1735 | C=0.8265 |
| 1000Genomes_30x | American | Sub | 980 | T=0.357 | C=0.643 |
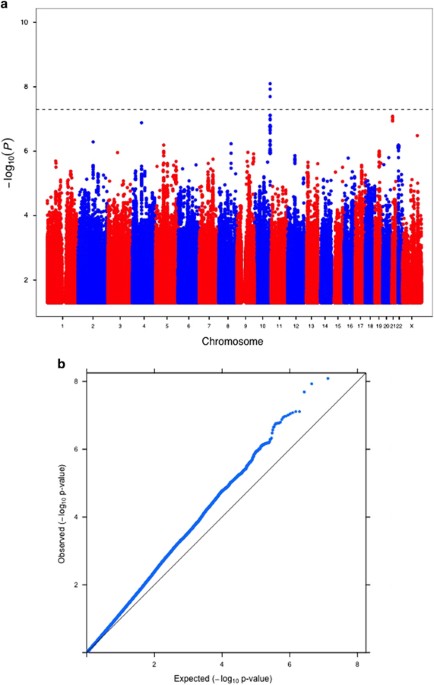
Polymorphisms (SNPs) in the CHRM2 gene on 7q31-35. From all individuals, standardized intelligence measures were available. Using a test of within-family association, which controls for the possible effects of population stratification, a highly significant association was found between the CHRM2 gene and intelligence. The strongest association was between rs324650 and performance IQ (PIQ), where the T allele was associated with an increase of 4.6 PIQ points. In parallel with a large family-based association, we observed an attenuated - although still significant - population-based association, illustrating that population stratification may decrease our chances of detecting allele-trait associations. Such a mechanism has been predicted earlier, and this article is one of the first to empirically show that family-based association methods are not only needed to guard against false positives, but are also invaluable in guarding against false negatives.

| 1000Genomes_30x | Global | Study-wide | 6404 | G=0.8482 | A=0.1518 |
| 1000Genomes_30x | African | Sub | 1786 | G=0.9675 | A=0.0325 |
| 1000Genomes_30x | Europe | Sub | 1266 | G=0.9171 | A=0.0829 |
| 1000Genomes_30x | South Asian | Sub | 1202 | G=0.7421 | A=0.2579 |
| 1000Genomes_30x | East Asian | Sub | 1170 | G=0.7085 | A=0.2915 |
| 1000Genomes_30x | American | Sub | 980 | G=0.839 | A=0.161 |





