Transcended Trucel
Peace & Dharma ; Vishwaguru India!
★★★★★
- Joined
- Feb 16, 2019
- Posts
- 48,630
My garbage genes are turning me mentally ill. only a bullet to the head can free meI hate my low genes
My garbage genes are turning me mentally ill. only a bullet to the head can free meI hate my low genes
Brutal, rope is always nearMy garbage genes are turning me mentally ill. only a bullet to the head can free me
This forum is turning my brain into a mush.My garbage genes are turning me mentally ill. only a bullet to the head can free me
my brain has turned to shit. I haven't talked to anyone IRL outside of family and cashiers in months. I am living in limbo. the days pass and I don't know anything. there are no new memories. when I look back the past year, there is nothing but infinite pain, not a single positive memory .This forum is turning my brain into a mush.
We report on the association of KIBRA with memory in two samples of older individuals assessed on either memory for semantically unrelated word stimuli (Rey Auditory Verbal Learning Test, n=2091), or a measure of semantically related material (the WAIS Logical Memory Test of prose-passage recall, n=542). SNP rs17070145 was associated with delayed recall of semantically unrelated items, but not with immediate recall for these stimuli, nor with either immediate or delayed recall for semantically related material. The pattern of results suggests a role for the T-->C substitution in intron 9 of KIBRA in a component of episodic memory involved in long-term storage but independent of processes shared with immediate recall such as rehearsal involved in acquisition and rehearsal or processes.

Kibra is a synaptic scaffold protein regulating learning and memory. Alterations of Kibra-encoding gene WWC1 cause various neuronal disorders, including Alzheimer's disease and Tourette syndrome. However, the molecular mechanism underlying Kibra's function in neurons is poorly understood. Here we discover that Kibra, via its N-terminal WW12 tandem domains, binds to a postsynaptic density enriched protein, Dendrin, with a nanomolar dissociation constant. On the basis of the structure of Kibra WW12 in complex with Dendrin PY motifs, we developed a potent peptide inhibitor capable of specifically blocking the binding between Kibra and Dendrin in neurons. Systematic administration of the inhibitory peptide attenuated excitatory synaptic transmission, completely blocked long-term potentiation induction, and impaired spatial learning and memory. A Kibra mutation found in Tourette syndrome patients causes defects in binding to Dendrin. Thus, Kibra can modulate spatial learning and memory via binding to Dendrin.

KIBRA has come into the focus of the neurogenetics field following the publication of human evidence pointing to an involvement of the gene in memory performance and cognition (Papassotiropoulos et al., 2006). In that publication, the authors report that carriers of the KIBRA/ WWC1 (rs17070145) T allele or, to a lesser extent, the calsyntenin 2 (CLSTN2) rs6439886 T allele performed significantly better on multiple episodic memory tasks than those homozygous for the C allele at either polymorphism. Furthermore, using functional magnetic resonance imaging (fMRI), they observed that brain activation (measured as oxygen extraction from blood) in key areas associated with memory retrieval was significantly greater in a selection of 15 WWC1 (rs17070145) T-allele-noncarriers than in 15 T-allele-carriers during an episodic memory task. After this initial finding, a considerable number of studies examined the WWC1 polymorphism in different contexts of cognition and in different populations (summarized in Table Table11 and reviewed below).

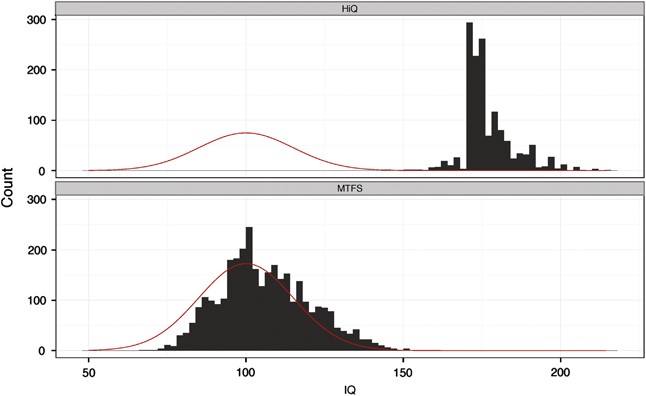
mogs me. I accept defeat. You are higher iq.
High IQ intellectuals cannot be differentiated from the regular layman in normal eyes, this is intentionally done to avoid the ban process.
Exactly, upon gazing at our stunning intellect, they will resort to something as primitive-minded as a “ban” To prevent us from intellectually, spiritually, physically invigorating and imbuing this forum with our combined intellectual might.
Fucking brutal.
This is sui-fuel for ethnic cels like me. Why does the most important trait in this world have to be immutable
Two measures of Educational Attainment (EA) were defined in accordance with the 1997 International Standard Classification of Education (ISCED) of the United Nations Educational, Scientific and Cultural Organization (UNESCO). This classification transforms each country-specific educational system into seven internationally comparable categories of EA (14). In each study, EA of the subjects was first transformed into the appropriate ISCED level of the country. Thereafter the equivalent to US years of schooling was imputed, as described in Table S2. In some countries the measures did not differentiate between levels 5 and 6. In these cases everyone with a tertiary education was coded as ISCED 5, and 20 years of schooling was imputed instead of 19. The resulting continuous measure of EA as US-schooling-year equivalents is abbreviated as EduYears throughout the manuscript.
We also analyzed the binary outcome, College, which differentiates between individuals who hold a tertiary degree and those who do not. This binary variable was imputed taking the value 1 if the individual had completed a college degree (ISCED level 5 or above of the ISCED classification), and 0 if the individual had not completed a college degree (ISCED level 4 or below).
EduYears may provide more information about individual differences within a country, but College may be more comparable across countries. Nonetheless, the point biserial correlation between the two measures is relatively high, e.g., 0.82 (in the STR sample), 0.74 (RS-I), 0.88 (RS-II) and 0.91 (RS-III). Note, however, that the EduYears analysis focuses on the effects at the mean of the phenotype distribution, whereas the College analysis focuses on differences between the upper tail of the phenotype distribution and the remaining values. The study-specific phenotype measurements and distributions are summarized in Table S3. All studies used a self-report of educational attainment, except STR. In STR, official register-based results for educational attainment were available. The descriptive statistics for the basic study-specific age and birth years are provided in Table S4. The combined discovery sample comprises 101,069 individuals for EduYears and 95,427 individuals for College. Analyses were performed at the cohort level according to a pre-specified analysis plan, which restricted
Mfw too low iq to understand the spreadsheet above
Mfw too low iq to understand the spreadsheet above
| Population | Group | Sample Size | Ref Allele | Alt Allele |
|---|
| Population | Group | Sample Size | Ref Allele | Alt Allele |
|---|---|---|---|---|
| Total | Global | 7920 | G=0.3301 | A=0.0000, C=0.6699, T=0.0000 |
| European | Sub | 7116 | G=0.2919 | A=0.0000, C=0.7081, T=0.0000 |
| African | Sub | 510 | G=0.820 | A=0.000, C=0.180, T=0.000 |
| African Others | Sub | 22 | G=0.77 | A=0.00, C=0.23, T=0.00 |
| African American | Sub | 488 | G=0.822 | A=0.000, C=0.178, T=0.000 |
| Asian | Sub | 4 | G=0.0 | A=0.0, C=1.0, T=0.0 |
| East Asian | Sub | 2 | G=0.0 | A=0.0, C=1.0, T=0.0 |
| Other Asian | Sub | 2 | G=0.0 | A=0.0, C=1.0, T=0.0 |
Genome-wide association meta-analysis of 78,308 individuals
identifies new loci and genes influencing human intelligence

1000Genomes Global Study-wide 5008 G=0.5895 A=0.4105 1000Genomes African Sub 1322 G=0.7118 A=0.2882 1000Genomes East Asian Sub 1008 G=0.5764 A=0.4236 1000Genomes Europe Sub 1006 G=0.4473 A=0.5527 1000Genomes South Asian Sub 978 G=0.603 A=0.397 1000Genomes American Sub 694 G=0.562 A=0.438 1000Genomes_30x Global Study-wide 6404 G=0.5934 A=0.4066 1000Genomes_30x African Sub 1786 G=0.7116 A=0.2884 1000Genomes_30x Europe Sub 1266 G=0.4542 A=0.5458 1000Genomes_30x South Asian Sub 1202 G=0.5957 A=0.4043 1000Genomes_30x East Asian Sub 1170 G=0.5803 A=0.4197 1000Genomes_30x American Sub 980 G=0.570 A=0.430 14KJPN JAPANESE Study-wide 28258 G=0.52371 A=0.47629 8.3KJPN JAPANESE Study-wide 16760 G=0.52243 A=0.47757 A Vietnamese Genetic Variation Database Global Study-wide 214 G=0.607 A=0.393 Allele Frequency Aggregator Total Global 10618 G=0.59314 A=0.40686, T=0.00000 Allele Frequency Aggregator European Sub 8276 G=0.5168 A=0.4832, T=0.0000
We identify common genetic variants associated with cognitive performance using a two-stage approach, which we call the proxy-phenotype method. First, we conduct a genome-wide association study of educational attainment in a large sample (n = 106,736), which produces a set of 69 education-associated SNPs. Second, using independent samples (n = 24,189), we measure the association of these education-associated SNPs with cognitive performance.
Three SNPs (rs1487441, rs7923609, and rs2721173) are significantly associated with cognitive performance after correction for multiple hypothesis testing
| Population | Group | Sample Size | Ref Allele | Alt Allele |
|---|
| Population | Group | Sample Size | Ref Allele | Alt Allele |
|---|---|---|---|---|
| Total | Global | 54534 | C=0.54260 | T=0.45740 |
| European | Sub | 35404 | C=0.52254 | T=0.47746 |
| African | Sub | 8228 | C=0.7058 | T=0.2942 |
| African Others | Sub | 276 | C=0.757 | T=0.243 |
| African American | Sub | 7952 | C=0.7040 | T=0.2960 |
| Asian | Sub | 484 | C=0.531 | T=0.469 |
| East Asian | Sub | 386 | C=0.565 | T=0.435 |
| Population | Group | Sample Size | Ref Allele | Alt Allele |
|---|
| Population | Group | Sample Size | Ref Allele | Alt Allele |
|---|---|---|---|---|
| Total | Global | 326666 | A=0.524533 | G=0.475467 |
| European | Sub | 285818 | A=0.511780 | G=0.488220 |
| African | Sub | 11002 | A=0.67424 | G=0.32576 |
| African Others | Sub | 402 | A=0.701 | G=0.299 |
| African American | Sub | 10600 | A=0.67321 | G=0.32679 |
| Asian | Sub | 3938 | A=0.6361 | G=0.3639 |
| East Asian | Sub | 3192 | A=0.6206 | G=0.3794 |
same. I've failed so many top tier job interviews that I want to ropeI legit can cope with being ugly but I can't fucking cope with being a low IQ retard it's actually so fucking brutal because I place a huge emphasis on obtaining and retaining knowledge. Its so fucking ovER I wish i could be an ugly Einstein
true, don't even bother about that genetic dna test or whatever the fuck these cumskins drag on our faces. It's a meme at this point just cope and push through in life.Being a curry nigger is really death sentence, there is no fucking DNA research. I cant even find any data on being currynigger. I wish I was a nigger or cumskin at the least, there would be genetic data to compare personal DNA to. This fucking world is truly horrible. There is no data on anything that fucking matters. Only speculations. What a shithole world. I would rather live in a world with fixed destiny and predetermined job,family, savings, death even if it was a bad life cause at the least their wouldnt be all this fucking volatiliy and change.
| Population | Group | Sample Size | Ref Allele | Alt Allele |
|---|
| Population | Group | Sample Size | Ref Allele | Alt Allele |
|---|---|---|---|---|
| Total | Global | 302410 | A=0.696548 | G=0.303452 |
| European | Sub | 267482 | A=0.676262 | G=0.323738 |
| African | Sub | 8382 | A=0.8890 | G=0.1110 |
| African Others | Sub | 318 | A=0.921 | G=0.079 |
| African American | Sub | 8064 | A=0.8878 | G=0.1122 |
| Asian | Sub | 3940 | A=0.9985 | G=0.0015 |
| East Asian | Sub | 3194 | A=0.9984 | G=0.0016 |
| Other Asian | Sub | 746 | A=0.999 | G=0.001 |
| Latin American 1 | Sub | 1134 | A=0.7399 | G=0.2601 |
| Latin American 2 | Sub | 7228 | A=0.8329 | G=0.1671 |
its over for us.
How do u even get a dna test? I really want one
A large body of twin and DNA-based research robustly demonstrates that AA and learning abilities are heritable. A recent meta-analysis of 61 twin studies reported a heritability estimate of 66% for AA. To date, GWAS of math ability using general population samples have been limited in size (N = 602 to 3000 individuals) and thus far have failed to identify robustly associated variants. In addition, there have been no published GWAS for standardized attainment scores in English and math during adolescence and no GWAS at all for science. The following study aims to assess whether academic subject-specific genetic contributions to English, math, and science exist.
Researchers performed genome-wide association studies of standardized national English, math, and science tests using the UK-based Avon Longitudinal Study of Parents and Children (ALSPAC) data. The performance in the three academic subjects were assessed using National Curriculum-based Standardized Assessment tests (SATs) at 11 and 14 years of age. The academic attainment average scores for English (N = 5983), math (N = 6017) and science (N = 6089) were calculated by summing age- and SAT scores from these two-time points for each academic subject. After the genotyping, one genome-wide significant single nucleotide polymorphism (SNP) was identified for attainment in science, but none for attainment in English or math. Further 26 independent SNPs showed suggestive evidence of association with science, 38 for math and 16 for English. The A allele of rs11264236 and the T allele of rs10905791 showed tendencies to increase academic scores in science. It also found that science was significantly more correlated with English and math than with each other, suggesting that academic performance in science might incorporate variance from the other two subjects.
While no significant differences in genetic correlations between academic subjects were found, the opposite pattern of associations was observed, with English and math being the most highly genetically correlated. One possible explanation is that the factors contributing to performance between science and maths, and science and English, are under greater environmental influence than those contributing to the correlation between English and maths performance, which correlate more for genetic reasons. Note, it is possible that this is a specific effect of this period of development for children. Read more about the study here:
Brutal, rope is always near
Comprehensive genomic analyses associate UGT8 variants with musical ability in a Mongolian population.
Park H et al. 2012
To identify genetic loci and variants that contribute to musical ability determined using a pitch-production accuracy test, researchers conducted family-based linkage and association analyses, and incorporated the results with data from exome sequencing and array comparative genomic hybridization analyses. They identified an intergenic single nucleotide polymorphism (SNP) near UGT8, a gene highly expressed in the central nervous system and known to act in brain organization. In addition, a non-synonymous SNP in UGT8 was revealed to be highly associated with musical ability.
jmg.bmj.com/content/49/12/747
| 1000Genomes_30x | Global | Study-wide | 6404 | T=0.7567 | C=0.2433 |
| 1000Genomes_30x | African | Sub | 1786 | T=0.6439 | C=0.3561 |
| 1000Genomes_30x | Europe | Sub | 1266 | T=0.6880 | C=0.3120 |
| 1000Genomes_30x | South Asian | Sub | 1202 | T=0.8469 | C=0.1531 |
| 1000Genomes_30x | East Asian | Sub | 1170 | T=0.9171 | C=0.0829 |
| 1000Genomes_30x | American | Sub | 980 | T=0.749 | C=0.251 |
Reference papers and your DNA
A genome-wide association study (GWAS) of the personality constructs in CPAI-2 in Taiwanese Hakka populations.
Kao PY et al. 2023
Researchers performed a genome-wide association studies (GWAS) to investigate the genetic components of the personality constructs in the Chinese Personality Assessment Inventory 2 (CPAI-2) in Taiwanese Hakka populations, who are likely the descendants of a recent admixture between a group of Chinese immigrants with high emigration intention and a group of the Taiwanese aboriginal population generally without it. The CPAI-2 that were not previously observed using the traditional Big Five personality measures and this is the first study that investigates the biological basis of CPAI-2. This inventory consists of 22 normal personality scales and 3 validity scales. For this study, they used 19 personality scales including the Social Potency factors such as Diversity, Logical vs Affective Orientation and Enterprise, Dependability factors such as Responsibility and Family Orientation. The first significant result was rs56666, located between the UTRN gene and the EPM2A, that is strongly associated with the personality, Enterprise. The second significant result was rs1267992, located in gene NKAIN2, that is strongly associated with Diversity. The final significant result was rs12503435, located between the RAPGEF2 gene and the FSTL5 gene, that is strongly associated with Logical vs. Affective Orientation. The participants with CC genotypes were likely to have logical and analytic thinking and behavior, while AA genotype participants were likely to be more sentimental and showed intuitive orientation. The RAPGEF2 gene is up-regulated in Alzheimer’s disease patients’ hippocampus and may play roles in Aβ oligomer-induced synaptic and cognitive degeneration. FSTL5 is expressed in cortical neurons, specifically in the hippocampus CA3 region and the cerebellum granular cell layer, and is important in synaptic transmission and plasticity.

This study examined the association between size of the caudate nuclei and intelligence. Based on the central role of the caudate in learning, as well as neuroimaging studies linking greater caudate volume to better attentional function, verbal ability, and dopamine receptor availability, we hypothesized the existence of a positive association between intelligence and caudate volume in three large independent samples of healthy adults (total N = 517). Regression of IQ onto bilateral caudate volume controlling for age, sex, and total brain volume indicated a significant positive correlation between caudate volume and intelligence, with a comparable magnitude of effect across each of the three samples. No other subcortical structures were independently associated with IQ, suggesting a specific biological link between caudate morphology and intelligence. Hum Brain Mapp 36:1407–1416, 2015. © 2014 Wiley Periodicals, Inc.
Background Musical abilities such as recognising music and singing performance serve as means for communication and are instruments in sexual selection. Specific regions of the brain have been found to be activated by musical stimuli, but these have rarely been extended to the discovery of genes and molecules associated with musical ability.
Methods A total of 1008 individuals from 73 families were enrolled and a pitch-production accuracy test was applied to determine musical ability. To identify genetic loci and variants that contribute to musical ability, we conducted family-based linkage and association analyses, and incorporated the results with data from exome sequencing and array comparative genomic hybridisation analyses.
Results We found significant evidence of linkage at 4q23 with the nearest marker D4S2986 (LOD=3.1), whose supporting interval overlaps a previous study in Finnish families, and identified an intergenic single nucleotide polymorphism (SNP) (rs1251078, p=8.4×10−17) near UGT8, a gene highly expressed in the central nervous system and known to act in brain organisation. In addition, a non-synonymous SNP in UGT8 was revealed to be highly associated with musical ability (rs4148254, p=8.0×10−17), and a 6.2 kb copy number loss near UGT8 showed a plausible association with musical ability (p=2.9×10−6).
Conclusions This study provides new insight into the genetics of musical ability, exemplifying a methodology to assign functional significance to synonymous and non-coding alleles by integrating multiple experimental methods.
| 1000Genomes_30x | Global | Study-wide | 6404 | T=0.7567 | C=0.2433 |
| 1000Genomes_30x | African | Sub | 1786 | T=0.6439 | C=0.3561 |
| 1000Genomes_30x | Europe | Sub | 1266 | T=0.6880 | C=0.3120 |
| 1000Genomes_30x | South Asian | Sub | 1202 | T=0.8469 | C=0.1531 |
| 1000Genomes_30x | East Asian | Sub | 1170 | T=0.9171 | C=0.0829 |
| 1000Genomes_30x | American | Sub | 980 | T=0.749 | C=0.251 |
| 1000Genomes_30x | Global | Study-wide | 6404 | C=0.8887 | A=0.1113 |
| 1000Genomes_30x | African | Sub | 1786 | C=0.9619 | A=0.0381 |
| 1000Genomes_30x | Europe | Sub | 1266 | C=0.6967 | A=0.3033 |
| 1000Genomes_30x | South Asian | Sub | 1202 | C=0.9426 | A=0.0574 |
| 1000Genomes_30x | East Asian | Sub | 1170 | C=0.9991 | A=0.0009 |
| 1000Genomes_30x | American | Sub | 980 | C=0.805 | A=0.195 |
| 1000Genomes_30x | Global | Study-wide | 6404 | C=0.4202 | T=0.5798 |
| 1000Genomes_30x | African | Sub | 1786 | C=0.7083 | T=0.2917 |
| 1000Genomes_30x | Europe | Sub | 1266 | C=0.3791 | T=0.6209 |
| 1000Genomes_30x | South Asian | Sub | 1202 | C=0.3436 | T=0.6564 |
| 1000Genomes_30x | East Asian | Sub | 1170 | C=0.2077 | T=0.7923 |
| 1000Genomes_30x | American | Sub | 980 | C=0.296 | T=0.704 |





